一、研究方向
针对我国天然橡胶供给严重不足和橡胶产业亟待升级的现实问题,围绕天然橡胶产量与质量形成的分子机制,开展橡胶树胶乳代谢调控尤其是乳管蔗糖代谢与胶乳再生的分子调控研究,以揭示影响胶乳再生能力和橡胶产量的分子调控机制,为我国天然橡胶产业发展和升级提供理论支持。
二、团队成员
现有在编科研人员6人,其中正高职称1人,副高职称4人,中级职称1人,具有海外科研经历2人,获海南省领军人才称号2人,获海南省拔尖人才称号3人,获海南省“南海名家—青年”培育计划2人,获热带农业青年英才—拔尖人才1人。 课题组长期聘用人员1名,培养毕业研究生31人(博士生8人)。

三、研究任务
(1)乳管蔗糖代谢与胶乳再生调控机制研究
橡胶生物合成是在乳管细胞中以蔗糖为原料进行的,乳管蔗糖的降解利用为橡胶合成提供物质和能量基础。围绕碳源向乳管转运以及乳管内碳源分配利用,阐明相关途径关键节点和关键基因,揭示碳源分配利用的调控模式和调控网络,为高效橡胶生产提供理论支持,为橡胶遗传改良提供分子基础。
(2)橡胶树组学大数据分析
综合利用基因组学、转录组学、蛋白组学、代谢组学、调控组学等技术,对橡胶树产胶、抗性等特定生物性状或过程从基因、转录、蛋白和代谢水平进行全面的深入的阐释,为橡胶树分子生物学研究提供高效平台和持续支撑。
(3)橡胶产量分子标记的挖掘与利用
利用前期研究结果得到的关键基因、小分子化合物、代谢产物等,开展群体调查、组学检测和关联分析,进行目的基因和功能性分子标记开发,用于品种鉴定、产量早期预测、目标品系筛选等。
四、研究进展
(1)获得迄今为止国际上质量最好的橡胶树基因组序列,组装大小1.37Gb,鉴定编码蛋白基因43877个,发现橡胶合成相关基因,特别是REF/SRPP家族发生了显著扩增和功能分化。发现乙烯处理不能增强内源乙烯合成,但显著激活信号应答和传导刺激橡胶增产。研究揭示了橡胶高产的遗传线索及乙烯刺激产胶新机制。论文发表在《Natrue Plants》,为该杂志最高引用论文之一。
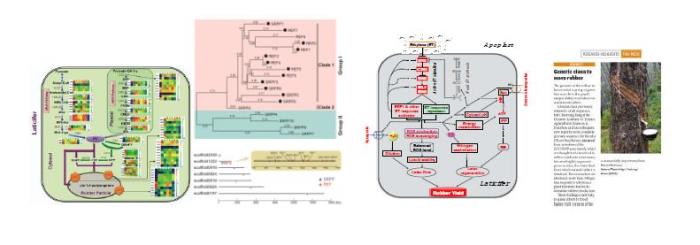
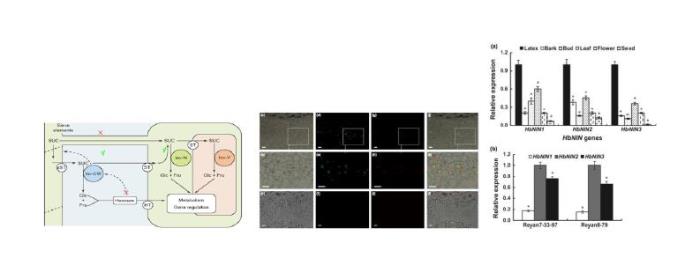
(2) 分离鉴定了决定乳管蔗糖供给关键基因HbSUT3,及降解关键基因HbNIN2,这些基因的表达和活性调控与橡胶产量密切相关。系统证实了乳管蔗糖代谢调控在橡胶产量形成中的核心地位,为橡胶高产遗传改良指明了重点方向。相关成果获得2017年海南省科技进步一等奖。
(3)丰富了橡胶树乳管蔗糖转运理论。发现定位于液泡的膜蛋白基因HbSUT 5在胶乳中仅次于HbSUT 3高表达,为典型的Suc-H+ 共转运体。在乙烯刺激和割胶处理实验中受诱导显著下调表达,与HbSUT 3的模式相反。乙烯处理下,胞质(胶乳)中蔗糖含量显著下降,而黄色体中的含量变化不大,表明SUT5的表达下降阻止了黄色体蔗糖进入胞质,从而加强了库组织蔗糖的饥渴度。揭示了乳管中HbSUT5和HbSUT3通过协同作用调控乳管蔗糖的供给,对胶乳代谢发挥重要调控作用。
(4)基于橡胶树的基因组,开展比较转录组学研究,确定乳管在基因数目及表达模式上均存在特异性;系统描述天然橡胶生物合成的代谢通路,进一步证实MVA途径为主要代谢通路,推测FPP为其合成起始物;明确泛素化修饰是乳管代谢的核心调控机制,且割胶促进乳管泛素化修饰,丰富了特异细胞—乳管的代谢机理。
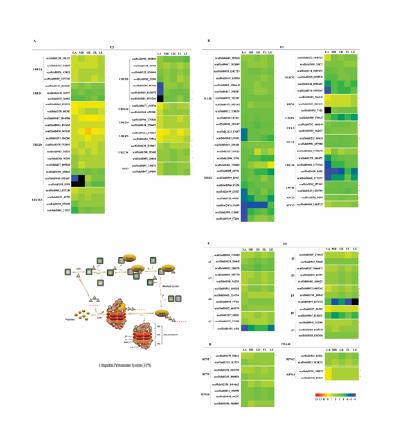
Terpenoid biosynthesis pathway and expression profiles. (A) MVA (mevalonate) pathway for rubber and steroid synthesis in the cytosol. Note: Intermediates in MVA related pathway: HMG-CoA, hydroxymethylglutaryl-CoA; MVA, mevalonate; MVA-P, mevalonate-5-phosphate; MVA-PP, mevalonate pyrophosphate; IPP, isopentenyl pyrophosphate; DMAPP, dimethylallyl pyrophosphate; FPP, farnesyl pyrophosphate; Enzymes in MVA related pathway: AACT, Acetyl-CoA C-acetyltransferase; HMGS, HMG-CoA synthase; HMGCR, HMG-CoA reductase; MVK, mevalonate kinase; PMVK, phosphomevalonate kinase; MVD, mevalonate pyrophosphate decarboxylase; IDI, IPP isomerase; FPPS, FPP synthase; SS, squalene synthase; CPT, cis-prenyltransferase; CBP, CPT binding protein; REF, rubber elongation factor protein; SRPP, small rubber particle protein. (B) MEP (2-C-methyl-D-erythritol-4-phosphat) pathway for carotenoid biosynthesis in the plastid. Note: Intermediates in MEP related pathway: Pyr, pyruvic acid; G3P, glyceraldehyde-3-phosphate; DOXP, 1-deoxy-D-xylulose-5-phosphate; MEP, 2-C-methyl-D-erythritol-4-phosphate; CDP-ME, 4-(cytidine-5′-diphospho)-2-Cmethyl-D-erythritol; CDP-MEP, 2-phospho-4-(cytidine-5′-diphospho)-2-C-methyl-D-erythritol; ME-CPP, 2-C-methyl-D-erythritol-2,4-cyclodiphosphate; HMBPP, 4-hydroxy-3-methylbut-2-enyl diphosphate; IPP, isopentenyl pyrophosphate; DMAPP, dimethylallyl pyrophosphate. GPP, geranyl pyrophosphate; GGPP, geranylgeranyl pyrophosphate. Enzymes in MEP related pathway: DXS, DOXP synthase; DXR, DOXP reductoisomerase; CMS CDP-ME synthase; CDK, CDP-ME kinase; MDS, ME-CPP synthase; HDS, HMBPP synthase; HDR, HMBPP reductase; GPPS, geranyl pyrophosphate synthase; GGPPS, geranyl pyrophosphate synthase; PSY, phytoene synthase. Log2 (TPM) values were used by Heml1.0.3.7 software, and TPM values were the average values of three biological repeats. Asterisk, latex-specific genes; red, up-regulated genes; green, down-regulated genes.

Transcription factor analysis. (A) Number of transcription factors in the rubber tree. Note: up-regulated TFs in the latex; down-regulated TFs in the latex; Unchanged TFs in the latex. (B) Transcription factor families of the rubber tree. Note: Top ten transcription factor families. MYB, myeloblastosis; AP2/ERF, APETALA 2/ethylene-responsive element binding factor; bHLH, basic Helix-Loop-Helix; NAC, NAM, ATAF1/2 and CUC2; HB, homeobox; GRAS, GAI (gibberellic acid insensitive), RGA (repressor of GAI-3 mutant) and SCR (scarecrow). (C) KEGG enrichment analysis of laticifer specific transcription factors. Note: up-regulated TFs in laticifer; down-regulated TFs in laticifer. (D) Expression profiles of MYC2, EIN3, and ERF1. Note: MYC, myelocytomatosis; EIN, ethylene insensitive; ERF, ethylene-responsive factor. Log2 (TPM) values were used by Heml1.0.3.7 software, and TPM values were the average values of three biological repeats. Asterisk, latex-specific genes; red, up-regulated genes; green, down-regulated genes.
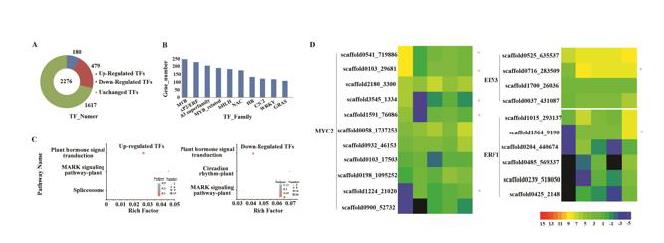
Ubiquitin-proteasome system and expression profiles. (A) Ubiquitin-conjugating enzymes (E2s); (B) Ubiquitin ligases (E3s); (C) 20S core particles of proteasome; (D) 19S regulatory particles of proteasome. Note: E2s: UBCE2, ubiquitin-conjugating enzyme 2; HIP2, uomeodomain-interacring protein kinase 2. E3s: SIAH1, seven in absentia homolog 1; PIRH2, p53 induced RING-H2 protein; MGRN1, mahogunin ring finger-1; RBX1, ring-box protein 1; CUL3, cullin 3; DCAF, DDB1-Cul4 associated factor; CDC20, cell division cycle protein 20; APC, anaphase promoting complex. 20S: α, alpha-subunit; β, beta-subunit. 19S-Lid: RPN, regulatory particles-no ATP ase. Log2 (TPM) values were used by Heml1.0.3.7 software, and TPM values were the average values of three biological repeats. Asterisk, latex-specific genes; red, up-regulated genes; green, down-regulated genes.
五、承担(参与)的在研省部级科研项目
课题组近五年获资助的省部级科研项目10余项,包括国家自然科学基金面上项目、国家重点研发专项子课题、国家留学基金及海南省基金等,部分项目如下:
1.“橡胶树乳管柠檬酸代谢在天然橡胶合成中的作用机制研究” 国家基金面上项目,201901-202212
2.“橡胶树乳管代谢调控转录因子的深度发掘与功能分析”国家基金面上项目,201801-202112
3.“橡胶树橡胶烃合成关键酶—ATP柠檬酸裂解酶基因的分离及功能分析”国家基金面上项目,201801-202112
4. “橡胶树乳管蔗糖降解关键转化酶HbNIN2的互作蛋白筛选与功能分析”国家基金面上项目,201601-201912
5. “割胶促进橡胶树产胶的分子机制研究”海南省创新团队,201701-201912
6. “天然橡胶产量形成与调控(参与)” 国家重点研发计划,201807-202212
7. “热带作物种质资源精准评价与基因发掘(参与)” 国家重点研发计划,201905-202212
8.“橡胶树乳管细胞无氧呼吸代谢关键酶PDC和LDH的调控机制”海南省基金人才项目,202001-202212
9.“橡胶树HbCIPK和HbCBL的互作及生物学功能研究” 海南省基金人才项目,202012-202312
六、主要研究成果
课题组成立以来,获海南省科技进步一等奖一项,获国家级科研项目资助19项,授权发明专利13项,发表论文70余篇,其中SCI论文20余篇。

代表性论文[1-13]
1. Long, X., et al., Latex-specific transcriptome analysis reveals mechanisms for latex metabolism and natural rubber biosynthesis in laticifers of Hevea brasiliensis. Industrial Crops and Products, 2021. 171.
2. Fang, P., et al., The hexokinases HbHXK2 and 4 are key enzymes involved in glucose metabolism and contribute to rubber productivity in Hevea brasiliensis (para rubber tree). Industrial crops and products, 2021. 159: p. 113025-.
3. Fang, P., et al., A predominant isoform of fructokinase, HbFRK2, is involved in Hevea brasiliensis (para rubber tree) latex yield and regeneration. Plant Physiology and Biochemistry, 2021. 162(6): p. 211-220.
4. Long, X., et al., Identification and evaluation of suitable reference genes for gene expression analysis in rubber tree leaf. Mol Biol Rep, 2020. 47(3): p. 1921-1933.
5. Long, X., et al., Characterization of a vacuolar sucrose transporter, HbSUT5, from Hevea brasiliensis: involvement in latex production through regulation of intracellular sucrose transport in the bark and laticifers. BMC Plant Biol, 2019. 19(1): p. 591.
6. Zhu, J., et al., Characterization of Sugar Contents and Sucrose Metabolizing Enzymes in Developing Leaves of Hevea brasiliensis. Front Plant Sci, 2018. 9: p. 58.
7. Huang, Y., et al., Characterization of the rubber tree metallothionein family reveals a role in mitigating the effects of reactive oxygen species associated with physiological stress. Tree Physiol, 2018. 38(6): p. 911-924.
8. Zhou, B., et al., Expressional characterization of two class I trehalose-6-phosphate synthase genes in Hevea brasiliensis (para rubber tree) suggests a role in rubber production. New Forests, 2017: p. 1-14.
9. Xiao, X.H., et al., The calcium-dependent protein kinase (CDPK) and CDPK-related kinase gene families in Hevea brasiliensis-comparison with five other plant species in structure, evolution, and expression. FEBS Open Bio, 2017. 7(1): p. 4-24.
10. Sui, J.L., et al., The SWEET gene family in Hevea brasiliensis - its evolution and expression compared with four other plant species. FEBS Open Bio, 2017. 7(12): p. 1943-1959.
11. Tang, C., et al., The rubber tree genome reveals new insights into rubber production and species adaptation. Nat Plants, 2016. 2(6): p. 16073.
12. Long, X., et al., Identification and Characterization of the Glucose-6-Phosphate Dehydrogenase Gene Family in the Para Rubber Tree, Hevea brasiliensis. Front Plant Sci, 2016. 7: p. 215.
13. Fang, Y., et al., De novo Transcriptome Analysis Reveals Distinct Defense Mechanisms by Young and Mature Leaves of Hevea brasiliensis (Para Rubber Tree). Sci Rep, 2016. 6: p. 33151.






